MicroWorldOmics: Data analysis desktop app for microbiome and virome
With the production of a large number of high-throughput sequencing data in the fields of ecology, medicine and pharmacy, the complexity of data analysis and interpretation is increasing. However, the microbiome and virome field still lacks a convenient and programming-free desktop application for comprehensive analysis of microbiome and virome data, especially in the context of virome analysis and "dark matter" exploration. Therefore, a plugin development model of desktop service MicroWorldOmics is proposed to provide a convenient one-stop desktop analysis application for life science and biomedical fields.
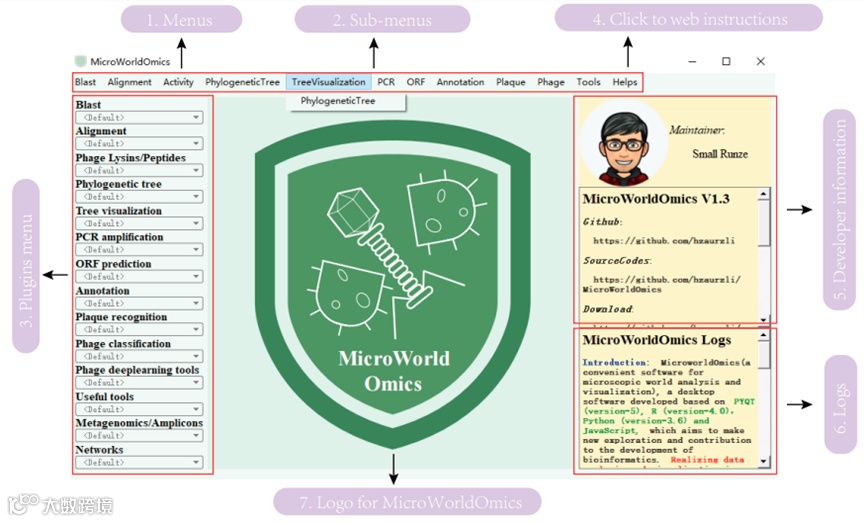
Figure1 MicroWorldOmics home page and its functions
In order to meet the above requirements, Li Runze from HZAU and Dong Wei from Sun Yat-sen University recently launched MicroWorldOmics, a desktop application that integrates microbiome and virome multi-omics analysis. The app related articles recently published in BioRxiv (https://doi.org/10.1101/2024.06.24.600528). And provided the open community (https://github.com/hzaurzli/MicroWorldOmics), the user can timely feedback problems.

Figure2 bioRxiv
MicroWorldOmics has the following functional modules:
Figure3 MicroWorldOmics main structure and design concept
1. Microbiome analysis:
(1) Metagenome: diversity analysis, abundance difference analysis, microbial marker identification, dimensionality reduction analysis, etc
(2) Network construction: Bayesian network construction, cosine similarity network construction, differential network analysis, etc
2. Virome analysis:
(1) Bacteriophage identification
(2) Determination of phage lysogenicity
(3) Identification of phage species
(4) phage host prediction
3. Exploring Dark matter:
(1) Analysis of endolysin activity
(2) Analysis of antimicrobial peptide activity
4. Epidemiological analysis:
(1) MLST analysis
(2) Serotype analysis
(3) Identification and analysis of virulence genes and drug resistance genes
Demo of MicroWorldOmics
In addition, MicroWorldOmics integrates many deep learning modules to assist in the analysis of relevant data, which makes some deep learning models developed based on Linux platforms can also run on Windows platforms, greatly improving the efficiency of experimenters in data analysis.
On the other hand, for some software that can only be compiled in Linux, we also rewrite it and make them run in the Windows system. If some software cannot be compiled on Windows, we also provide remote servers, users only need to upload files in the corresponding plug-in to the remote server for running, waiting for the data to download to the client.

Figure4 Plugin development concept
MicroWorldOmics has established more than 90 subapplications, and more than 600 R packages and Python modules have been called. Among them, 7 subprograms have been developed independently, and the remaining subprograms have been developed from public software repositories (Github, Biopython, Bioconductor, etc). One of the features of MicroWorldOmics when used cross-platform is not only the cross-operating system (Windows, Linux, Mac users can use), but also the comprehensive use of cross-language software (including software based on Python, R, Cpp languages). MicroWorldOmics uses a plugin development model that allows for parallel computing of multiple plugins if computing resources allow.
In addition, each application provides sample data (about 90 personalization features in total), and data entry prompts are displayed on the page of each plugin for user convenience.
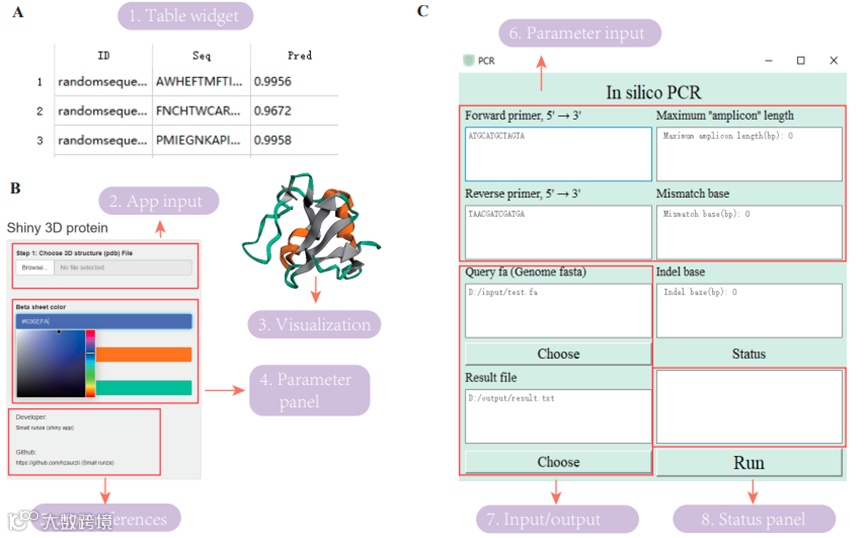
Figure5 Plugin parameter example interface
All of the analysis features in MicroWorldOmics support custom grouping and parameter Settings, and the results are presented in high quality charts, suitable for publication, and available for free download. In addition, the results table has filtering and sorting functions for users to filter and explore.
After continuous optimization, MicroWorldOmics V1.3 has been launched, a good software needs to be constantly maintained and updated. We sincerely invite all users in the process of use, if you find any problems or have any suggestions, welcome to give us feedback. We will seriously consider these valuable suggestions and make improvements to contribute to the construction of domestic bioinformatics software. Contact information is (Email: yun_act@163.com), thank you for your support and cooperation!



